GraphAssembly#
GraphAssembly provides graph-based representation and analysis of CAD assemblies. Nodes represent shapes (solids), and edges capture relationships like distance, contact, and geometric constraints.
Note
Prerequisites: Graph Fundamentals
GraphAssembly#
User documentation for the GraphAssembly class of the graph.assembly module of the volmdlr_tools library
What is it?#
GraphAssembly is a class from a submodule of the volmdlr_tools library. The goal of this module is to create graphs from a geometric structure and to use it a base structure to:
Add metadata to the CAD
On the edge linking CAD elements
On the nodes representing CAD elements
Extract sub-graph / specific nodes / specific edges base on filters
Save already computed information on the graph in a serializable object
The class GraphAssembly is:
as generic as possible
provide base methods to manipulate is
inherits from the class
Graphand specify some CAD-assembly dedicated methods
Base functionalities#
Custom typings#
NodeID = int
Attributes = dict[str, Any]
Node = tuple[NodeID, Attributes]
Edge = tuple[NodeID, NodeID]
Class attributes#
graph: networkx.GraphThe network graph of the assemblyroot_compound: CompoundThe root compound of the assembly, optional.
Properties#
nodes: networkx.NodeViewA getter of the nodes of the graph that provide access to their attributes. Usedata=Trueargument to get aNodeDataViewto also get the attributes.edges: networkx.EdgeViewA getter of the edges of the graph that provide access to their attributes. Usedata=Trueargument to get aEdgeDataViewto also get the attributes.
Instantiation#
A GraphAssembly object can instantiate from a VolumeModel or from a Compound
For now, it is made to work with VolumeModel with an unique Compound as a root primitive, which are the type of VolumeModel we have when load a CAD file (STEP or native geometry).
# From a volmdlr.model.VolumeModel
@classmethod
def from_volume_model(cls, volume_model: VolumeModel) -> "GraphAssembly":
# From a volmdlr.shapes.Compound
@classmethod
def from_compound(cls, compound: Compound) -> "GraphAssembly":
# From a networkx.Graph
@classmethod
def from_networkx_graph(cls, graph: nx.Graph) -> "Graph":
Default nodes attributes#
At the instantiation with from_volume_model or from_compound class methods, the nodes have default attributes:
name: The name of the corresponding geometry elements (extracted from the VolumeModel)level: The deep level of the corresponding geometry element (in the assembly tree)class: The class of the corresponding geometry element.color: The color of the corresponding geometry element, in a plot_data way.shape: The shape used for plotting with plot_data.primitive_id: The ID of the corresponding geometry element, to retrieve the geometry from the node.location: The location (translation and rotation) of the corresponding geometry element.
Basic getters#
Several methods are made to extract information from the GraphAssemby object
# Get the number of nodes of the graph.
@property
def n_nodes(self) -> int:
# Get the number of edges of the graph.
@property
def n_edges(self) -> int:
# Create the networkx graph from the object attributes.
def network_graph(self) -> nx.Graph:
# Get a specific attribute of a specific Node.
def get_node_attribute(self, node: NodeID, attribute: str) -> Any:
# Get a specific attribute of a specific Edge.
def get_edge_attribute(self, edge: Edge, attribute: str) -> Any:
# Get the Shape (i.e. the geometry) that corresponds to a specific node.
def get_shape(self, node_id: NodeID) -> Shape:
Also, the __getitem__ method is set to access the node or edge attributes:
def __getitem__(self, item: Union[NodeID, Edge]) -> Attributes:
# Example usage
graph: GraphAssembly # graph is a GraphAssembly instance
# NODE
graph[1] # Return all the attributes of node with id 1
graph.nodes[1] # Equivalent
graph[1]["name"] # Return the value of attribute "name" of the node with id 1
graph.get_node_attribute(1, "name") # Equivalent, except there is a None default
# value if attribute does not exist
# EDGE
graph[(1, 2)] # Return all the attribute of the Edge between node 1 and 2
graph.edges[(1, 2)] # Equivalent
graph[(1, 2)]["distance"] # Return the value of attribute "distance" of the edge (1, 2)
graph.get_edge_attribute((1, 2), "distance") # Equivalent, but with None default value
Plotting#
The graph can be plotted with plot data
def plot_data(self, reference_path: str = "#", **kwargs) -> list[plot_graph.NetworkxGraph]:
# Example usage
graph: GraphAssembly # graph is a GraphAssembly instance
graph.plot()
Example plot
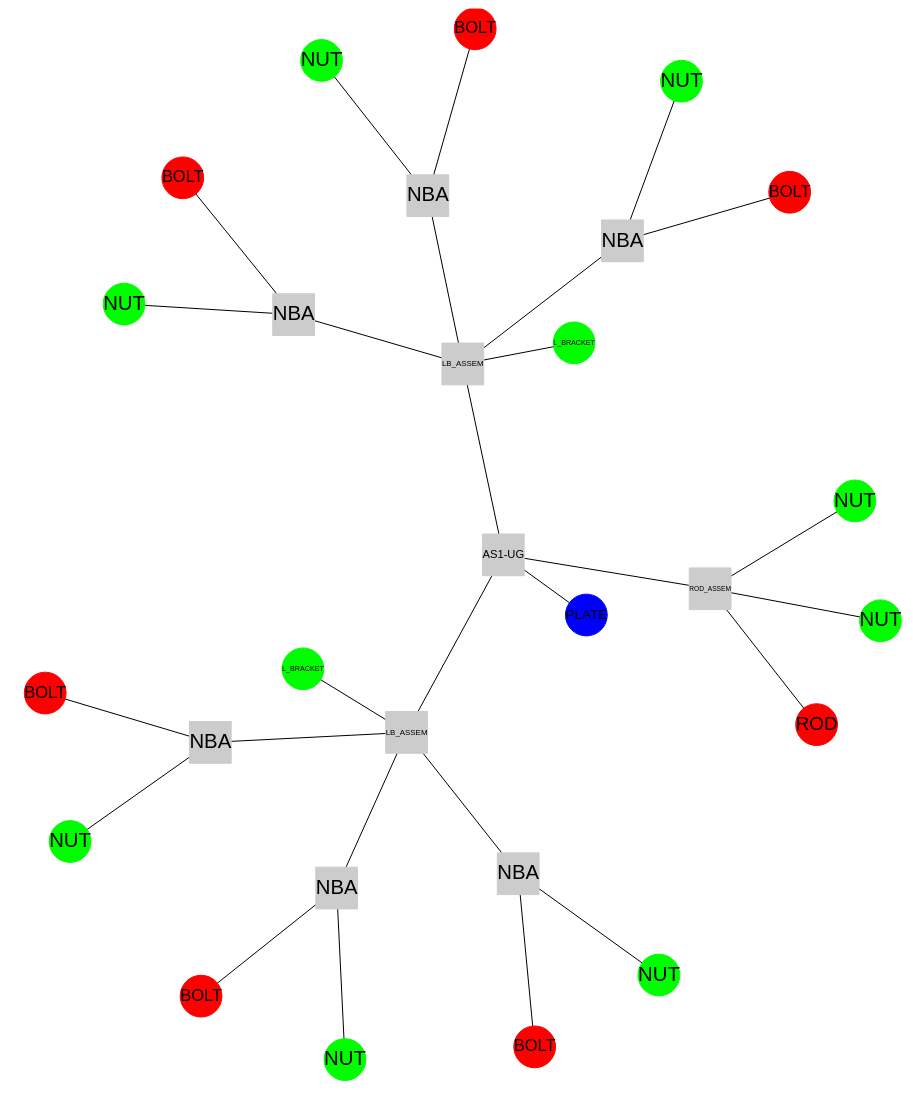
Corresponds to this
VolumeModel: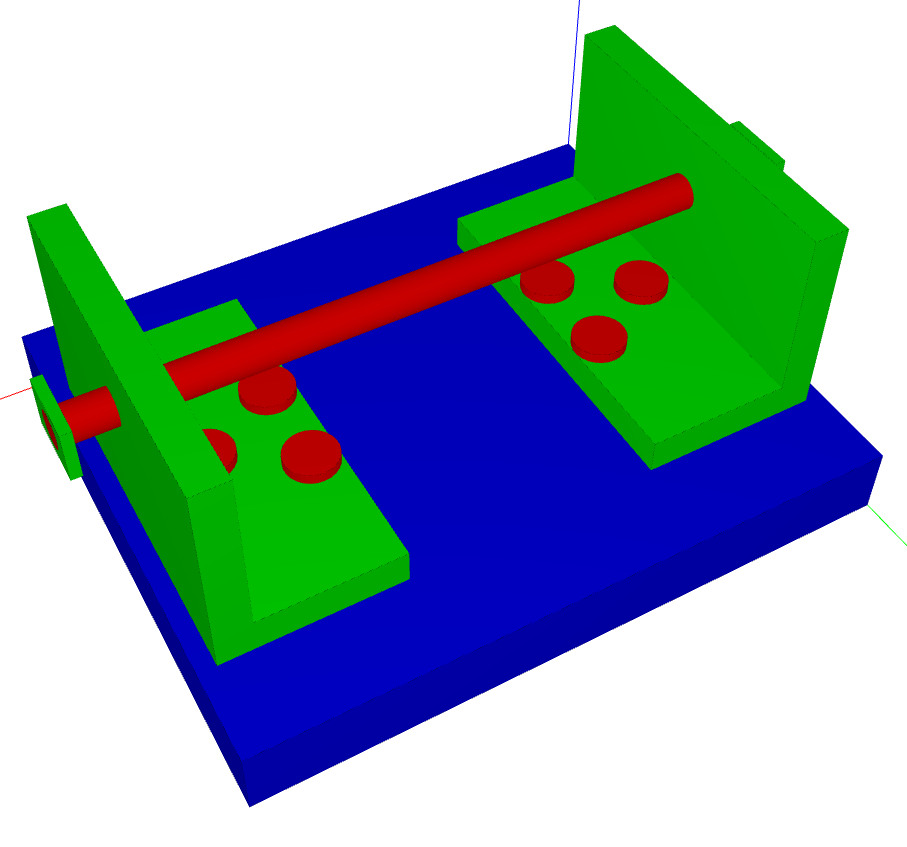
Basic setters#
Like basic getters, several methods allow setting attribute on specific node / edge:
# Set a specific attribute on a specific node.
def set_node_attribute(self, node: NodeID, attribute: str, value: Any) -> None:
# Set a specific attribute on a specific edge.
def set_edge_attribute(self, edge: Edge, attribute: str, value: Any) -> None:
# Create an edge with given attributes, if not already existing.
def create_edge(self, edge: Edge, attributes: Attributes = None) -> None:
Advanced functionalities#
We have seen how to get or set attributes on specific nodes / edges directly by passing the value. Now we will see how to:
set attributes dynamically computed on specific filtered nodes / edges
Example: set the distance between all solids of the graph
extract specific nodes / edges based on a given filter
Example: extract all the solids at less than 10mm from a specific solid of the graph
The is made possible by passing callable as methods arguments.
These have to be methods or functions that take a NodeID or an Edge (depending on the case) as argument and that compute a value (for attribute setting) or a bool (for filtering).
The typing is always specified (expected argument of the callable and expected return type) in methods that expect such argument.
The different typings are:
# A callable that take a NodeID and return a value (for attribute setting).
Callable[[NodeID], Any]
# A callable that take an Edge and return a value (for attribute setting).
Callable[[Edge], Any]
# A callable that take a NodeID and return a bool (for filtering).
Callable[[NodeID], bool]
# A callable that take an Edge and return a bool (for filtering).
Callable[[Edge], bool]
Advanced getters#
These getters can extract edges / nodes based on a filtering function.
# Get all the nodes that satisfy the filter_function (all by default)
def get_nodes(
self, filter_function: Callable[[NodeID], bool] = lambda node: True
) -> NodesMap:
# Get all the edges that satisfy the filter_function (all by default)
def get_edges(
self, filter_function: Callable[[Edge], bool] = lambda edge: True
) -> EdgesMap:
Advanced setters#
These setters can set dynamically computed attribute on filtered edges or nodes
# Set a dynamically computed attribute on all the nodes that satisfy the filter_function
def set_nodes_attribute(
self,
attribute: str,
attribute_function: Callable[[NodeID], Any],
filter_function: Optional[Callable[[NodeID], bool]] = None,
) -> None:
# Set a dynamically computed attribute on all the edges that satisfy the filter_function
def set_edges_attribute(
self,
attribute: str,
attribute_function: Callable[[Edge], Any],
filter_function: Optional[Callable[[Edge], bool]] = None,
) -> None:
# Create an edge between all the node that satisfy a filter_function
def create_edges(self, filter_function: Optional[Callable[[Edge], bool]]) -> None:
Creating edge is mandatory because it is not possible to set attributes on node that does not exist.
Extract#
Some methods allow extracting data a different way from getters methods
# Extract the neighbors of given node
def neighbors(
self, node: NodeID, filter_function: Callable[[Edge], bool] = lambda edge: True
) -> NodesMap:
# Extract a subgraph with all the node that fulfill the filter function
def subgraph(self, filter_function: Callable[[NodeID], bool]) -> "Graph":
Specific methods for common usage#
Some specific methods have been implemented:
For commonly use metadata
As good example of the class usage
def set_bounding_box_attribute(self) -> None:
"""Set the bounding box attribute for all nodes."""
self.set_nodes_attribute(
"bounding_box", lambda node: self.get_shape(node).bounding_box
)
def set_distance_attribute(self) -> None:
"""Set the distance attribute between all solids."""
# Create an edge between all solids
self.create_edges(
filter_function=lambda edge: self[edge[0]]["class"] == "Solid"
and self[edge[1]]["class"] == "Solid"
)
# Set the distance attribute
self.set_edges_attribute(
attribute="distance",
attribute_function=lambda edge: self.get_shape(edge[0]).distance(
self.get_shape(edge[1])
),
filter_function=lambda edge: self[edge[0]]["class"] == "Solid"
and self[edge[1]]["class"] == "Solid",
)
def set_connection_geometry(self, tol: float = 1e-9) -> None:
"""
Set the connection geometry attribute for all solids. The edge attribute "connection" will be set.
:param tol: The tolerance to consider two solids as connected.
"""
# Create an edge between all solids
self.create_edges(
filter_function=lambda edge: self[edge[0]]["class"] == "Solid"
and self[edge[1]]["class"] == "Solid"
)
# Set the distance attribute if not already set
self.set_edges_attribute(
attribute="distance",
attribute_function=lambda edge: self.get_shape(edge[0]).distance(
self.get_shape(edge[1])
),
filter_function=lambda edge: self[edge].get("distance") is None
and self[edge[0]]["class"] == "Solid"
and self[edge[1]]["class"] == "Solid",
)
def connection_geometry(edge: Edge) -> list[Shape]:
"""Compute the connection geometry between two solids."""
solid1 = self.get_shape(edge[0])
solid2 = self.get_shape(edge[1])
connections = []
for shell1 in solid1.primitives:
for shell2 in solid2.primitives:
connections.extend(shell1.intersection(shell2, tol=tol))
return connections
# Set the connection geometry attribute
self.set_edges_attribute(
attribute="connection",
attribute_function=connection_geometry,
filter_function=lambda edge: self[edge].get("distance", 1e9) <= tol
and self[edge[0]]["class"] == "Solid"
and self[edge[1]]["class"] == "Solid",
)
def get_close_solids(
self, solid_node_id: NodeID, maximum_distance: float
) -> NodesMap:
"""
Get the close solids from a solid.
:param solid_node_id: The node id of the solid.
:param maximum_distance: The maximum distance to consider a solid as close.
:return: A dict of the close solids with their attributes.
"""
# Check if it's a solid
if self[solid_node_id]["class"] != "Solid":
raise ValueError("Provided node id does not correspond to a Solid.")
# Create the edges between the solid and the other
self.create_edges(
filter_function=lambda edge: solid_node_id in edge
and self[edge[0]]["class"] == "Solid"
and self[edge[1]]["class"] == "Solid"
)
# Compute the distance on these edges
self.set_edges_attribute(
attribute="distance",
attribute_function=lambda edge: self.get_shape(edge[0]).distance(
self.get_shape(edge[1])
),
filter_function=lambda edge: solid_node_id in edge
and self[edge[0]]["class"] == "Solid"
and self[edge[1]]["class"] == "Solid",
)
# Return the neighbors base on the distance
return self.neighbors(
solid_node_id,
lambda edge: self[edge].get("distance", 1e9) < maximum_distance,
)
Rules check examples#
Some scripts in the library shows how to use the class to check specific rules, on the following geometry:

1_position_rule.py: Check that all the nuts are under the plate2_distance_rule.py: Check if the bolts are at a minimum distance between each other3_connection_rule.py: Check that the rod is only connected to other solid with “Cylindrical” connection4_proximity_rule.py: The ROD should not have more than 4 solids at a given distance, and the plate should not be part of it
The key in checking rules is:
Having a way to identify the involved components
Having a way to compute a data that allow verifying the rule
See Also#
Graph Fundamentals - Base graph concepts
Attributed Adjacency Graph (AAG) - Face-level adjacency graphs
Kinematics & Determinacy - Kinematic analysis using GraphAssembly